The anatomy of the neural network allows the brain to function properly. Confocal microscopy and subsequent analysis of the imaged data is a way to understand the anatomy and physiology of neural circuits and relationships within neural networks. Neurons as specific filamentary structures can be analyzed manually or automatically using specialized software tools.
Matthias G. Haberl and colleagues have addressed this issue and developed a new procedure for fluorescent labeling and automatic analysis of voluminous microscope data.
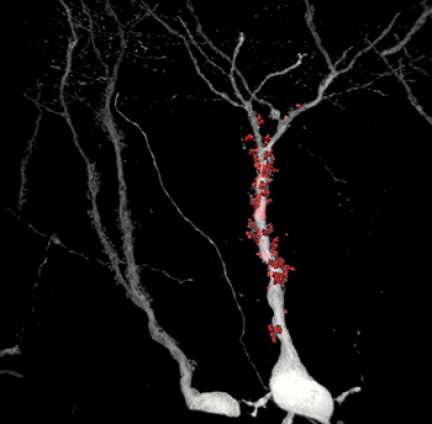
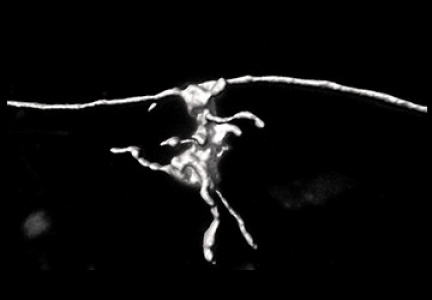
Figure 1: The high S/N ratio data obtained using the viral(rebies) vector has enabled scientists to accurately reconstruct these structures in 3D - part of specific synapses.
The neurons are labeled with a viral vector that is expressed as an intense fluorescent protein in the cells, which then fills all neuronal protrusions, including very long axons. The samples thus prepared are imaged using a confocal microscope and then analysed. Using dedicated software, a 3D model of whole dendrites, dendritic spines and synapses is created. These models are then used to quantify key anatomical parameters.
In contrast to most conventional approaches, neurons of physiologically older model organisms can be studied using the "Haberl method". This allowed the researchers to reconstruct the fine morphological details of the mouse brain of an elderly individual with induced Alzheimer's disease. This method provides new avenues for research into this widespread neurodegenerative disease.
Imaris software with the FilamentTracer module offers a very intuitive and flexible setup for analyzing filamentary structures (neurons, filamentous proteins, blood vessels):
- 3D visualization and filament analysis
- Detection and tracking of filamentary structures
- Algorithms for automatic analysis
- Possibility of manual processing or manual finishing
Webinar
Introduction To Imaris For Neuroscientists - Imaris 8.4
"Large-scale analysis of 3D neuron morphology" Accessed July 10, 2018 www.bitplane.com.